|
1 Shemyakin Institute of Bioorganic chemistry, USSR Academy of Sciences.
Cytokines make up a family of glycoprotein growth factors that have been shown to support clonal proliferation of hematopoietic progenitor cells [1]. Interleukin-3 (IL-3) and granulocytemacrophage colony-stimulating factor (CSF-2) stimulate differentiation and proliferation of cell progenitors along multiple (myeloid and erythroid) pathways [2-4]. Interleukin-4 (IL-4) can affect B cell, T cell, mast cell and macrophage functions [5]. Interleukin-5 (IL-5) is a more specific cytokine, acting on later stages of eosinophil differentiation [6]. The human genes for CSF -2, IL-3, IL4, and IL-5 have been cloned, sequenced [3, 7-9], and mapped by in situ hybridization to human chromosome 5 at bands q23-31 [10-15], a region that is frequently deleted in patients with myeloid disorders [del(5q)] and acute myeloid leukemia (AML) [16, 17]. A close genomic linkage of human IL-3 and CSF-2 genes was reported [18, 19]. The distance between the genes was found to be only 10.5 kilobases (kb). The physical linkage of the IL-4 and IL-5 genes to within 240310 kb was also demonstrated by longrange mapping, using pulse-field gel electrophoresis (PFGE) [15, 20]. Close linkage between the IL-3 and CSF-2 genes and between IL-4 and IL-5 genes, together with the similar gene structure, regulation, and biological activities of the four genes [21 ], suggests that they may have been derived from a common ancestral gene(s) and might be apart of a gene cluster related to the putative antioncogene that is involved in the development of AML and therapy-related acute nonlymphocytic leukemia. We report the molecular cloning and characterization of three regions of the long arm of human chromosome 5 that contain the CSF-2, IL-3, IL-4, and IL-5 genes; we also studied the physical organization of these genes using PFGE and hybridization probes derived from chromosome walking.
To isolate genomic DNA clones containing the genes for human CSF-2,
IL-3, IL4, and IL-5, phage and cosmid genomic libraries of 1.5 x
10 high6 and 2 x 10 high 6 clones, respectively, were prepared from
human leukocyte DNA. The libraries were probed with synthetic oligonucleotides
from the published sequences. Three cosmids and eight phage clones
identified with the IL-3 and CSF-2 probes at the first step of the
genomic walk cover 70 kb (Fig. 1) [22]. To continue the cosmid walk
in both directions, we set out to isolate single copy fragments
devoid of repetitive sequences from both extremities of the 70-kb
region. We subcloned the end fragments of cosmids cos-2 and cos-C
by digestion with restriction enzymes that have a unique site in
the polylinker region of the cosmid vector , followed by circularization
[23]. On the basis of primary structure of the insert DNA of these
clones, oligonucleotide 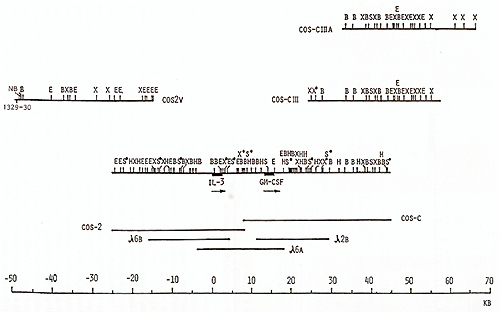 Fig. 1. Maps of the cloned 70-kb region that includes the genes for IL-3 and CSF-2 and cosmid clones that represent a 125-kb region: E, Eco RI; X, Xba I; B, Bam HI; H, Hin d III; S, Sal I; S*, X*, Xho I; N, NotI; Sf, Sfi I; B*, BssHII. Cosmid and phage overlap. Localization of oligonucleotide probe 1329-30 is shown 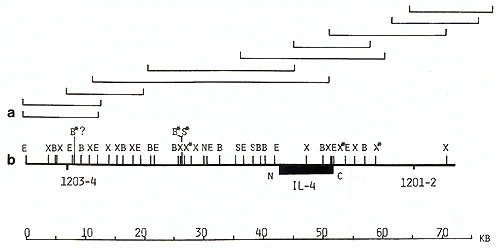 Fig. 2 a, b. Map of the cloned 75-kb region that includes the IL-4 gene: E, Ecol; X, Xbal; B, BamHI; X*, Xhol; S, Sfil; B*, BssHII; S*, Sac II. Cosmid and phage overlap. Localization of oligonucleotide probes 1201-2 and 1203-4 is shown 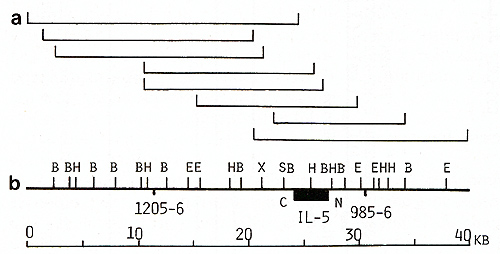
probes were synthesized. They were used to hybridize the cosmid
library, yielding cosmids cos-2V, cos-CIII, and cos-ClIIA (Fig.1).
In total, the cosmid and phage walk comprised at least eight overlapping
clones covering a 125-kb region around the IL-3 and CSF-2 genes.
Analogous genomic walks were performed around the IL-4 and IL-5
genes. More then six overlapping phage and cosmid clones represented
the 75-kb IL-4 gene region (Fig. 2). A 40-kb region of the IL-5
gene was covered by two cosmid and at least three phage clones (Fig.
3). Fur ther walking in both directions from the IL-5 gene was blocked
by the presence of repetitive sequences. We chose to analyze the
sites for the rare-cutting restriction enzymes in three cloned regions.
These enzymes usually have one or two CpGs in their recognition
sequences and are methylation sensitive. The dinucleotide CpG, which
is frequently methylated in a tissue-specific fashion, is underrepresent
in bulk mammalian DNA by a factor of 5. Unmethylated restriction
sites for the rare-cutting enzymes are often found clustered in
"CpG-rich islands," GC-rich regions of 1- 2 kb where CpG is found
at close to the frequency expected from local base composition and
where methylation is suppressed. These islands are frequently found
at, and mark, the 5' ends of genes [24-26]. We mapped BssHII, NolI,
SfiI, and Sac II restriction sites to trace possible CpG-rich islands.
As shown in Figs. 1 and 2, within the IL-3-CSF-2 region and the
IL-4 region we found single restriction sites for B.ssHII and SacII.
Only one NolI restriction site was present around 50 kb upstream
from the IL-3-CSF-2 gene cluster (Fig. 1 ); this CpG-rich island
may correspond to an unidentified gene because it lies adjacent
to unique sequences conserved in evolution (data not shown). To
generate a large-scale map of the regions around the IL-3-CSF-2
and IL4- IL-5 gene clusters we used rare-cutting restriction enzymes
and PFGE. Several probes from each region derived from walking and
partial sequencing were used for hybridization (Figs. 1-3). The
results of several hybridization experiments are shown in Figs.4-6;
the sizes of the restriction fragments that hybridized to different
probes are given in Tables 1 and 2. Our data confirmed physical
linkage of the IL-4 and IL-5 genes shown earlier by hybridization
with other probes [15, 20]: Bss HII, SfiI, and Nae I digests revealed
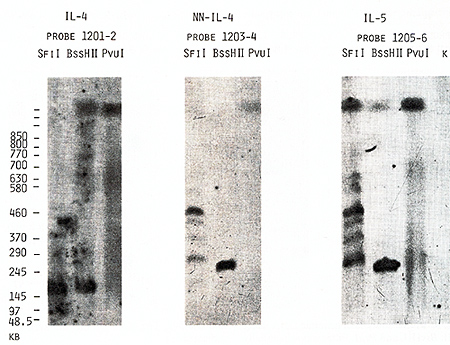
the same bands with probes 1203-4 and 1205-6 (Figs. 4,5). Based
on the length of the B,s.s HII and Sac II fragments (Table 2), we
estimated the distance between the two genes as 240 kb. Genomic
walks performed in the regions of the IL-4 and IL-5 genes allowed
us to determine the orientation of both genes by using for hybridization
the probes specific for the 5' and 3' ends of the genes separately
(Figs.4-6; Tables 1, 2). The results of these experiments allowed
us to place the 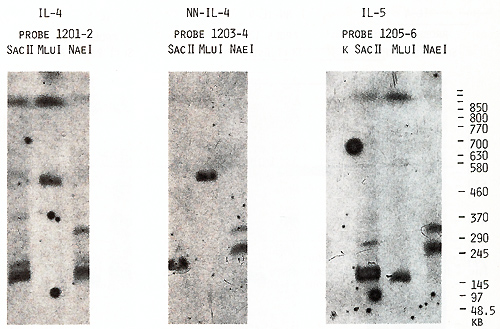 Fig.5. PFGE analysis of genomic DNA using Sfil, BssHll, and Pvul. Filter was sequentially hybridized to probes 1201-2, 1203-4, and 1205-6. The sizes of the markers are indicated 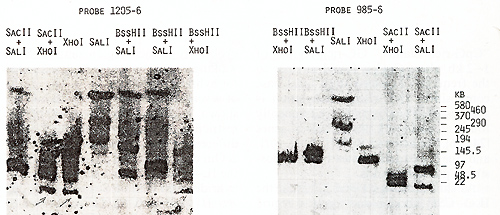 Fig.6. PFGE analysis of genomic DNA using SaIl, Xhol, Bs.sHII + Xhol, B.ssHll + SaIl, SacII +Xhol, SacII+SalI. Filter was hy bridized to probes 1205-6 and 985-6. The sizes of the markers are indicated genes on a long-range restriction map of this cluster in head to head orientation (Fig. 8). To test a possible linkage between the IL-4- IL-5 and IL-3-CSF-2 gene clusters we used for hybridization the probe 1329 30 mapped 50 kb upstreme from the 5'end of the IL-3 gene (Fig. 1). No common bands were detected in NotI digests with this probe and IL-4-specific probe 1201-2 (Fig. 7) and we were unable to demonstrate physical linkage of the two gene Table 1. Sizes in kilobase of restriction
fragments obtained from complete and partial digestions 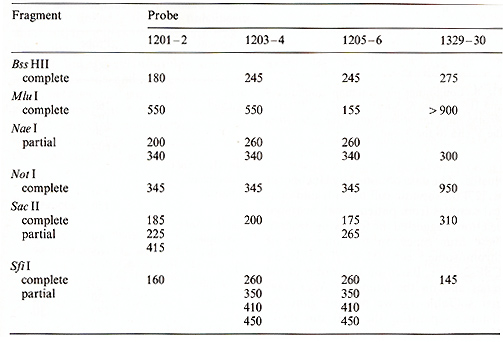 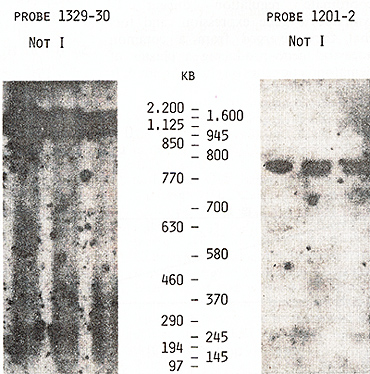 Fig. 7. PFGE analysis of genomic DNA using Not I. Filter was hybridized to probes 1329-30 and 1201-2. The sizes of the markers are indicated
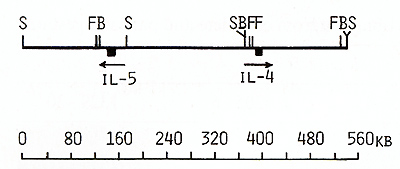 Fig. 8. Long-range physical map around IL-4 and IL-5 genes: S, Sac II; F, SfiI; B, Bss HII Arrows show the orientation of the genes from the 5' end to the 3' end clusters, The data obtained by Huebner et al, [27] on somatic cell hybrids and clinical samples from patients with acquired deletions suggested the following order of these four genes on the long arm of chromosome 5: cen-(IL-4-IL-5)-IL-3CSF-2-qter, Based on this order of the genes and on the length of NotI fragments (Table 1 ), we propose that the distance separating the IL-3 and IL-4IL-5 genes was not less than 1050 kb, Because of the close linkage of the four related genes, particularly the very close linkage of the IL-3 and CSF-2 genes, it might be suggested that they may have coordinate regulation during T lymphocyte gene expression, and (or) that they diverged from a common ancestral gene producing a cluster of hematopoietic genes on chromosome 5. Table 2. Sizes in kilobases of restriction
fragments obtained from complete and double digestions  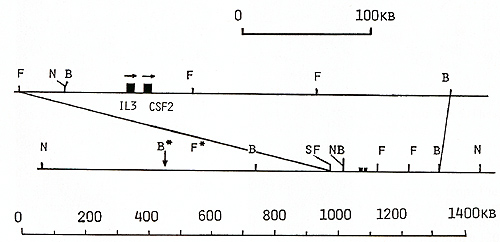
It was shown that in the mouse genome the IL-3 and CSF-2 genes are also closely physically linked they are only 14 kb apart and have the same orientation as in the human genome [15]. The conservation of linkage relationships supports both suggestions, and further study of the DNA between and surrounding the four genes is important in order to determine whether any other genes related to regulation of hematopoiesis are localized in the same cluster of genes.
1. Metcalf D (1984) The hematopoietic colony-stimulating factors. Elsevier, New York, Oxford, Chapter II. Tissue and cellular sources of the colony stimulation factors, p 309-329 2. IhIe lN, Keller l, Oroszlan S, Henderson lE, Copeland TD, Fitch F, Prystowsky MB, Ooldwasser E, Schrader lW, Palaszynski E, Dy M, lebel B (1983) Biologic properties of homogeneous interleukin 3. I. Demonstration of WEHI-3 growth factor activity, mast cell growth factor activity, P cell-stimulating factor activity, and hystamine-producing cell-stimulating factor activity. 1 Immunol131 :282-287 3. Yang Yu-Ch, Ciarletta AB, Temple PA, Chung MP, Kovacic S, Witek-Oianotti lS, leary AC, Kriz R, Donahue RE, Wong 00, Clark SC (1986) Human IL-3 (multiSCF) identification by expression cloning of a novel hematopoietic growth factor related to murine IL-3. Cell 47:3-10 4. Cantrell MA, Anderson D, Cerretti DP, Price V, McKeregham K, Tushinski Rl, Mochizuki DY, Karsen A, Orabstein K, Oillis S, Cosman D (1985) Cloning, sequence, and expression of a human granulocyte/macrophage colony-stimulating factor. Proc Natl Acad Sci USA 82:6250-6254 5. Yakota T, Otsuka T, Mosmann T, Banchereau l, Defrance T, Blanchard D, De Vries lE, Lee F, Arai K (1986) Isolation and characterization of a human interleukin cDNA clone, homologous to mouse B-cell stimulatory factor I, that expresses B-cell- and T -cell-stimulating activities. Proc Natl Acad Sci USA 83:5894-5898 6. Campbell HD, Tucker WQl, Hort Y, Martinson ME, Mayo O, Clutterbuck El, Sanderson Cl, Young IO (1987) Molecular cloning and expression of the gene encoding human eosinophil differentiation factor (interleukin-5). Proc Natl Acad Sci USA 84:6629-6633 7. Miyatake S, Otsuka T, Yokota T, lee F, Arai K (1985) Structure of the chromosomal gene for granulocyte-macrophage colony stimulating factor: Comparison of the mouse and human genes. EMBO 1 4:2561-256 8. Yokota T, Hagiwara H, Takebe Y, Otsuka T, Miyajima A, Meyerson P, Hoy P, Yokota K, Coffman R, Rennick D, Mos mann T, Howard M, Bancherau l, DeVries l, Arai N, Lee F, Arai K (1987) Isolation and characterization of mouse and human cDNA clones encoding IL-4 and IgA-enhancing factor/eosionophil CSF (IL-5). In: Webb DR, Pierce CW, Cohen S (eds) Molecular basis of Iymphokine action. Humana, Clifton, Nl, pp313-319 9. Tanabe T, Konishi M, Mizuta T, Noma T, Honjo T (1987) Molecular cloning and structure of the human interleukin-5 gene. 1 Bioi Cell 262:16580-16591 10. le Beau MM, Epstein ND, O'Brien Sl, Nienhuis A W, Yang Y -c, Clark SC, Rowley lD (1987) The interleukin 3 gene is located on human chromosome 5 and is deleted in myeloid leukemias with a deletion of 5q. Proc Natl Acad Sci USA 84:5913-5917 11. Huebner K, Isobe M, Croce CM, Oolde DW, Kaufman SE, Oasson 1 C (1985) The human gene encoding OM-CSF is at 5q21-32, the chromosome region deleted in the 5 q -anomaly. Science 230: 1282 1285 12. Sutherland OR, Baker E, Callen DF, Campbell HD, Young 10, Sanderson Cl, Lopez AF, Vadas MA (1988) Interleukin-5 is at 5q31 and is deleted in the 5 q -syndrome. Blood 71 : 1150-1152 13. Le Beau MM, Lemons RS, Espinosa RIII, Larson RA, Arai N, Rowley lD (1989) Interleukin-4 and interleukin-5 map to human chromosome 5 in a region encoding growth factors and receptors and are deleted in myeloid leukemias with a del(5q). Blood 73:647-650 14. Takahashi M, Yoshida MC, Satoh H, Hilders l, Yaoita Y, Honjo T (1989) Chromosomal mapping of the mouse Il-4 and human lL-5 genes. Genomics 4:47-52 15. Nienhuis WG, Bunn HF, Turner PH, Gopal TV, Nash WG, O'Brien SJ, Sherr CJ (1985) Expression of the human c-fms proto-oncogene in hematopoietic cells and its deletion in the 5 q -syndrome. Cell 42:421-428 16. Le Beau MM, Westbrook CA, Diaz MO, Larson RA, Rowley JD, Gasson JC, Golde DW, Sherr CJ (1986) Evidence of the involvement of GM-SCF and FMS genes in the deletion (5q) in the myeloid disorders. Science 231 : 984- 986 17. Yang Y-C, Kovacic S, Kriz R, Wolf S, Clark SC, Wellems TE, Nienhuis A, Epstein N (1988) The human genes for GM-CSF and lL-3 are closely linked in tandem on chromosome 5. Blood 71 :958-961 18. Ovchinnikov JuA, Dolganov GM, Frolova El (1988) Close linkage of the genes for lL-3 and GM-CSF in human genome. Proc Natl Acad Sci USSR 303:750-752 19. Chandrasekharappa SC, Rebelsky MS, Firak TA, Le Beau MM, Westbrook CA ( 1990) Along range restriction map of the interleukin-4 and interleukin-5 linkage group on chromosome 5. Genomics 6:94-99 20. Sideras P, Noma T, Honjo T (1988) Structure and function of interleukin 4 and 5. lmmunol Rev 102:189-212 21. Frolova El, Dolganov GM, Mazo lA, Smirnov DV, Copeland P, Stewart C, O'Brien SJ, Dean M (1991) Linkage mapping of the human CSF2 and IL3 genes. Proc Natl Acad Sci USA 88 :4821-4824 22. Heilig R, Lemaire C, Mandel JL (1987) A 230 kb cosmid walk in the Duchenne muscular dystrophy gene: detection of a conserved sequence and of a possible deletion prone region. Nucleic Acids Res 15 :9129-9142 23. Bird AP ( 1986) CpG-rich islands and the function of DNA methylation. Nature 321:209-213 24. Lindsay S, Bird AP (1987) Use of restriction enzymes to detect potential gene sequences in mammalian DNA. Nature 327:336-338 25. Gardiner-Garden M, Frommer M (1987) CpG islands in vertebrate genomes. J Mol BioI 196:261-282 26. Huebner K, Nagarajan L, Besa E, Angert E, Lange BJ, Cannizzaro LA, van den Berghe H, Santoli D, Finan J, Croce CM, Nowel PC (1990) Order of the genes on human chromosome 5q with respect to 5q interstitial deletions. Am J Hum Genet 46:26-36 |